Scientists played TIC-TAC-toe Board on the nanometer from the DNA
The match lasted six days. “TIC” won.
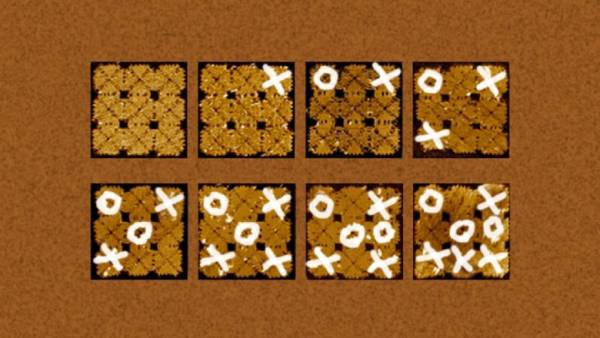 A team of biochemists from the California Institute of technology has continued to develop its approach to self-assembled DNA tiles according to the technology of DNA origami. Scientists have created a game Board for playing TIC-TAC-toe using the techniques of fractal Assembly. Noughts and crosses was also of DNA, was collected and was attached to the right cell of the playing Board with the help of technology DNA origami. The article describes one of the matches that researchers conducted in the lab — it lasted six days, “TIC” won.DNA origami is a technology build structures, and even mechanisms at the nanoscale. It is based on the fact that the DNA molecule has the property of “self-Assembly”.
A team of biochemists from the California Institute of technology has continued to develop its approach to self-assembled DNA tiles according to the technology of DNA origami. Scientists have created a game Board for playing TIC-TAC-toe using the techniques of fractal Assembly. Noughts and crosses was also of DNA, was collected and was attached to the right cell of the playing Board with the help of technology DNA origami. The article describes one of the matches that researchers conducted in the lab — it lasted six days, “TIC” won.DNA origami is a technology build structures, and even mechanisms at the nanoscale. It is based on the fact that the DNA molecule has the property of “self-Assembly”.
DNA strands consist of sahkopostin cores (actually threads) and attached the four types of molecules, called bases: adenine, guanine, cytosine and thymine, abbreviated with the letters a, G, C and T. the Feature of the reason that they have the property of complementarity, that is, can attract each other by hydrogen bonds, and in a specific combination. Adenine connects only with thymine, and guanine only with cytosine. Accordingly, any sequence of bases will seek to pair with complementary sequence. For example, ALGCA to connect with TEACHT.
If you take a small section of one DNA strand and connect it to complementary sites in the manner of a zipper, you get structure with “sticky” thread and a loop at the end, which biochemists call a “clip”. With the help of such clips placed in the right places larger DNA strands, you can get those to fold in complex two-dimensional structure. The Creator of the DNA origami Paul Rothmund were made of DNA molecules smiley faces, stars, and labels.

Very soon scientists learned how to do such DNA strands and three-dimensional structures, e.g., octahedrons, and even the simplest of nano-robots. A logical development of the idea was to suggest that separate DNA figures can be combined in a single macrostructure, creating repeating patterns. Managed a team of biochemists from the California Institute of technology under the guidance of Lulu Qian. They created the methodology of fractal Assembly.
This technique allows you to connect small items such DNA images in large two-dimensional arrays. In their work the authors proposed an algorithm, which allowed to determine the primary sequence of the DNA molecule and the “staples” that I fold it into the desired shape in order to obtain a predetermined image.
So scientists from Caltech assembled using DNA origami on a square element with sides less than 100 nanometers, consisting of 22 small DNA segments connected by “clips”.
In the right places to it was attached a label, which then can use atomic force microscope to see a pixel. The result is an image element, consisting of about 100 pixels.
Joining the tiles together via different types of “clips” scientists have received images of the Mona Lisa, cock, bacteria and Samatau boards in a square with sides of several hundred nanometers.
To collect image tiles for the tiles, it would take too many “staples” of different types, so scientists went the other way: they began to collect tiles step by step, how to collect first small region of the puzzle, and then over larger areas, and of those, finally, to create a complete puzzle. Every tile used the same four ribs, but as these puzzles are collected separately, there is no risk, for example, that the corner tile will be in the wrong corner.